Accelerating Drug Discovery: From FASTA Files to GenAI Insights on Databricks
How to build an end-to-end pipeline combining Data Engineering, Protein Language Models, and GenAI on the Databricks Platform.
Summary
- Process biological data at scale using Lakeflow Declarative Pipelines to transform raw FASTA protein sequences into analysis-ready tables in Unity Catalog.
- Classify proteins with transformer models by leveraging ProtBERT, a protein language model, to identify membrane transport proteins—key drug targets.
- Query protein insights in natural language through AI Functions that connect LLMs directly to your data, enabling researchers to explore promising drug candidates conversationally.
Drug development is notoriously slow and expensive. The average Research and Development (R&D) lifecycle spans 10-15 years, with a significant portion of candidates failing during clinical trials. A major bottleneck has been in identifying the right protein targets early in the process.
Proteins are the "working molecules" of living organisms—they catalyze reactions, transport molecules, and act as the targets for most modern drugs. The ability to rapidly classify proteins, understand their properties, and identify under-researched candidates could dramatically accelerate the discovery process (e.g. Wozniak et al., 2024, Nature Chemical Biology).
This is where the convergence of data engineering, machine learning (ML), and generative AI becomes transformative. In fact, you can build this entire pipeline on a single platform – the Databricks Data Intelligence Platform.
What We're Building
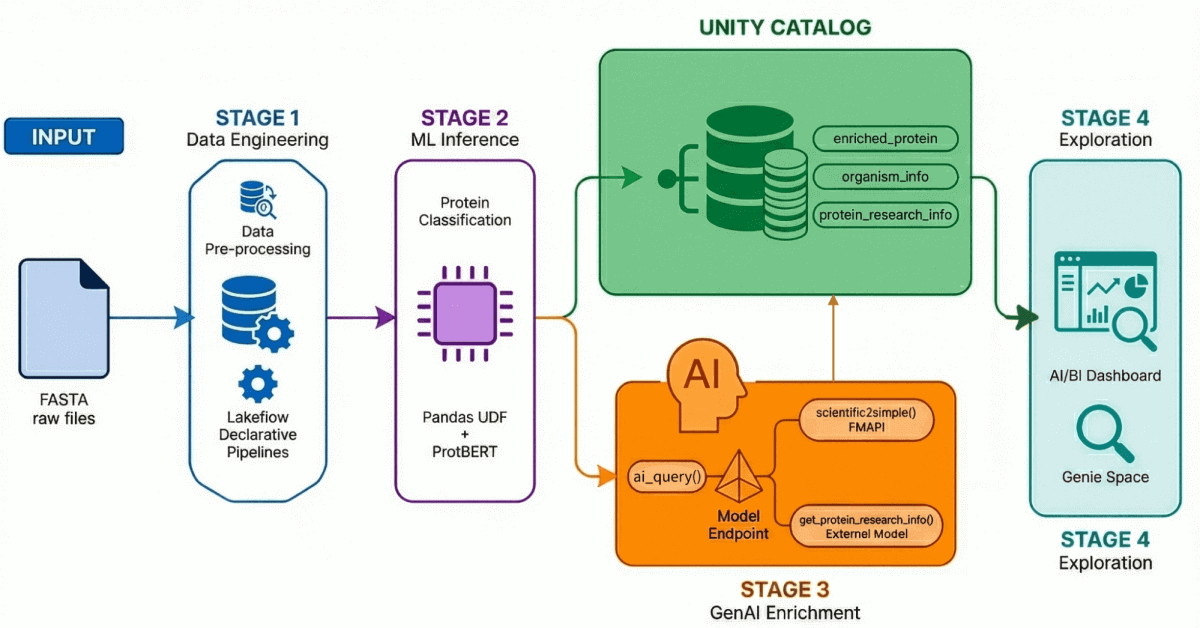
Our AI-Driven Drug Discovery Solution Accelerator demonstrates an end-to-end workflow through four key processes:
- Data Ingestion and Processing: Over 500,000 protein sequences are ingested and processed from UniProt.
- AI-Powered Classification: A transformer model is used to classify these proteins as either water-soluble or membrane transport.
- Insight Generation: Protein data is enriched with LLM-generated research insights.
- Natural Language Exploration: All processed and enriched data is made accessible through an AI-enabled dashboard and environment that supports natural language querying.
Let's walk through each stage:
Stage 1: Data Engineering with Lakeflow Declarative Pipelines
Raw biological data rarely arrives in a clean, analysis-ready format. Our source data comes as FASTA files—a standard format for representing protein sequences that looks something like this:
To the untrained eye, this sequence data is nearly impossible to interpret—a dense string of single-letter amino acid codes. Yet, by the end of this pipeline, researchers can query this same data in natural language, asking questions like "Show me under-researched membrane proteins in humans with high classification confidence" and receiving actionable insights in return.
Using Lakeflow Declarative Pipelines, we build a medallion architecture that progressively refines this data:
- Bronze Layer: Raw ingestion of FASTA files using BioPython, extracting IDs and sequences.
- Silver Layer: Parsing and structuring—we extract protein names, organism information, gene names, and other metadata using regex transformations.
- Gold/Enriched Layer: Curated, analysis-ready data enriched with derived metrics like molecular weight—ready for dashboards, ML models, and downstream research. This is the trusted layer that analysts and scientists query directly.
The result: Clean, governed protein data in Unity Catalog, ready for downstream ML and analytics. Critically, the data lineage that extends beyond this stage to the other stages (highlighted below) provides incredible value for scientific reproducibility.
Stage 2: Protein Classification with Transformer Models
Not all proteins are created equal when it comes to drug discovery. Membrane transport proteins—those embedded in cell membranes—are particularly important drug targets because they control what enters and exits cells.
We leverage ProtBERT-BFD, a BERT-based protein language model from the Rostlab, fine-tuned specifically for membrane protein classification. This model treats amino acid sequences like language, learning contextual relationships between residues to predict protein function.
The model outputs a classification (as Membrane or Soluble) along with a confidence score, which we write back to Unity Catalog for downstream filtering and analysis.
Stage 3: Enriching Data with GenAI
Classification tells us what a protein is. But researchers need to know why it matters—what is the recent research? Where are the gaps? Is this an under-explored drug target?
This is where we bring in LLMs. Leveraging both Databricks' Foundational Model API as well as External Model endpoints, we create registered AI Functions that enriches protein records with research context.
Stage 4: Natural Language Exploration
We bring everything together in an AI/BI Dashboard with Genie Space enabled.
Researchers can now:
- Filter proteins by organism, classification score, and protein type
- Explore distributions of molecular weights and classification confidence
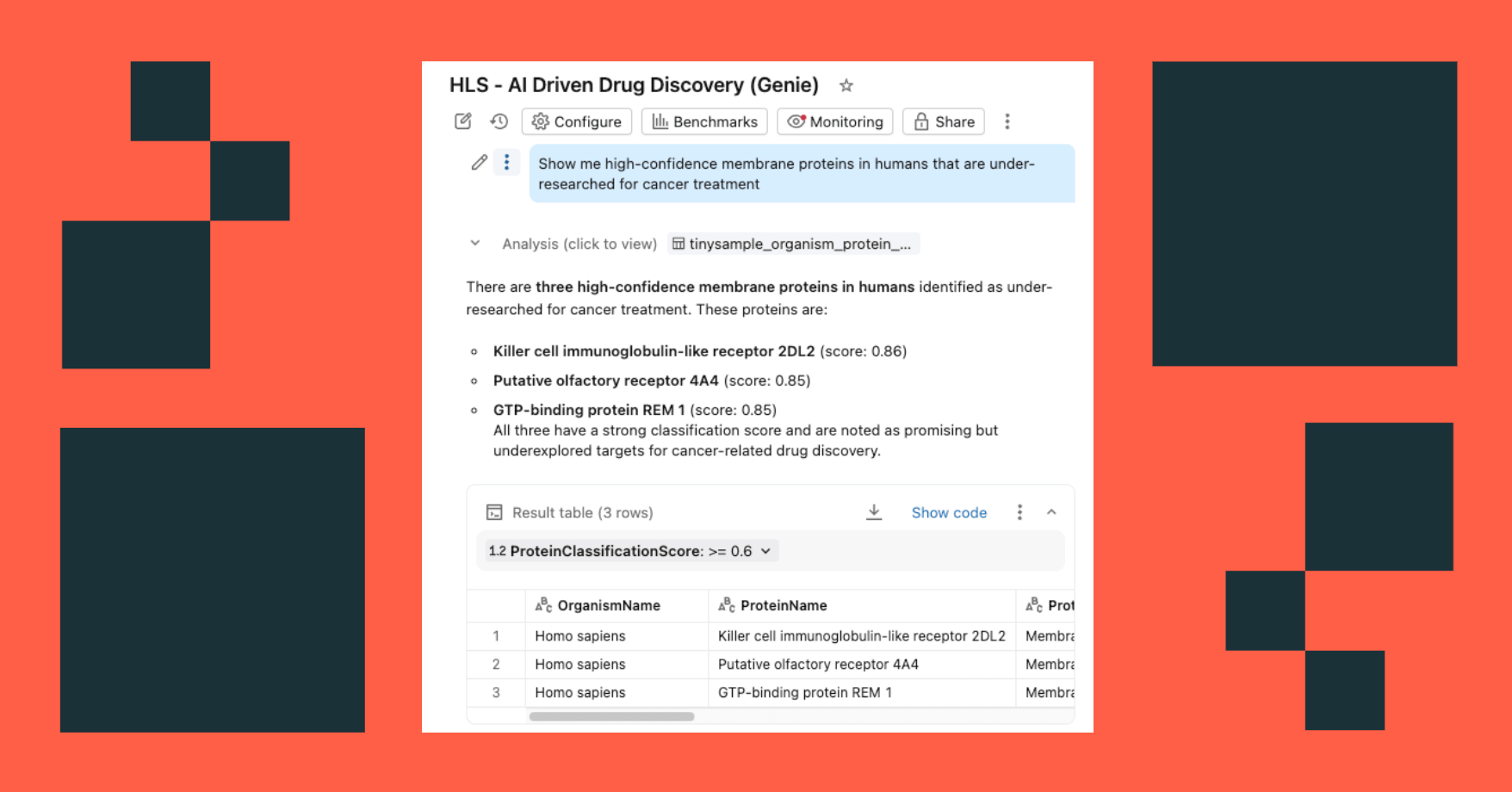
- Ask questions in natural language: "Show me high-confidence membrane proteins in humans that are under-researched for cancer treatment"
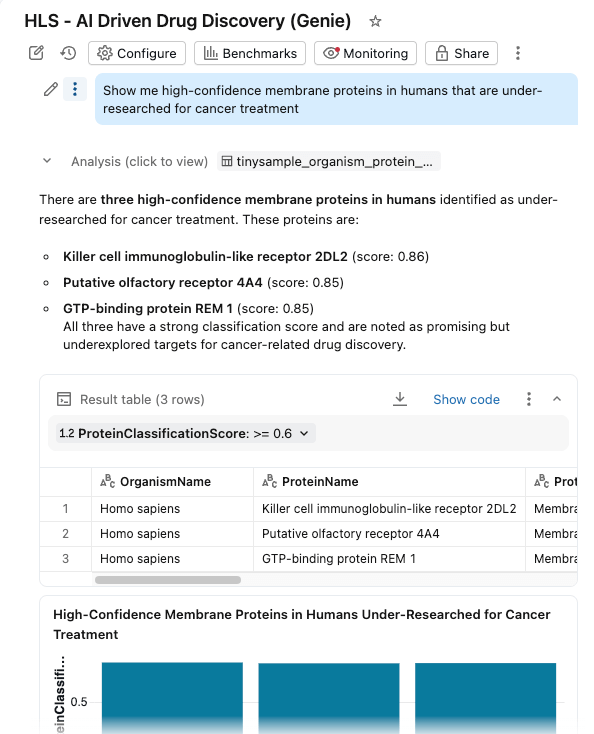
The dashboard queries the same governed tables in Unity Catalog, with AI Functions providing on-demand (or batch processed) enrichment.
Data intelligence reshapes industries
The Power of a Unified Platform
What makes this solution compelling is not due to any single component—it is that everything runs on one platform:
| Capability | Databricks Feature |
|---|---|
| Data Ingestion & ETL | Lakeflow Declarative Pipelines |
| Data Governance | Unity Catalog |
| ML inference | GPU Compute |
| LLM integration | FMAPI + External Models + AI Functions |
| Analytics | Databricks SQL |
| Exploration | AI/BI Dashboards + AI/BI Genie Space |
Critically, there is no data movement between systems. No separate MLOps infrastructure. No disconnected BI tools. The protein sequence that enters the pipeline flows through transformation, classification, enrichment, and ends up queryable in natural language—all within the same governed environment.
The complete solution accelerator is available on GitHub:
github.com/databricks-industry-solutions/ai-driven-drug-discovery
What's Next
This accelerator demonstrates the art of the possible. In production, you might extend it to:
- Process the full UniProt database with provisioned throughput endpoints
- Add more (open or custom) classification models for different protein properties
- Build RAG pipelines over scientific literature for more grounded LLM responses
- Integrate with downstream molecular simulation workflows
- Connect to protein structure prediction (AlphaFold/ESMFold) to add 3D structural context to classified proteins
- Extend to other genomic formats (FASTQ, VCF, BAM) using Glow for large-scale sequencing and variant analysis
The foundation is there. The platform is unified. The only limit is the science you want to accelerate. Get started today.
Never miss a Databricks post
Sign up
What's next?

Data Science and ML
November 14, 2024/2 min read
Providence Health: Scaling ML/AI Projects with Databricks Mosaic AI
Product
November 27, 2024/6 min read