De-identifying Medical Images Cost-Effectively with Vision Language Models on Databricks
VLMs are surprisingly good at de-identifying medical images and are cost-effective at Databricks.
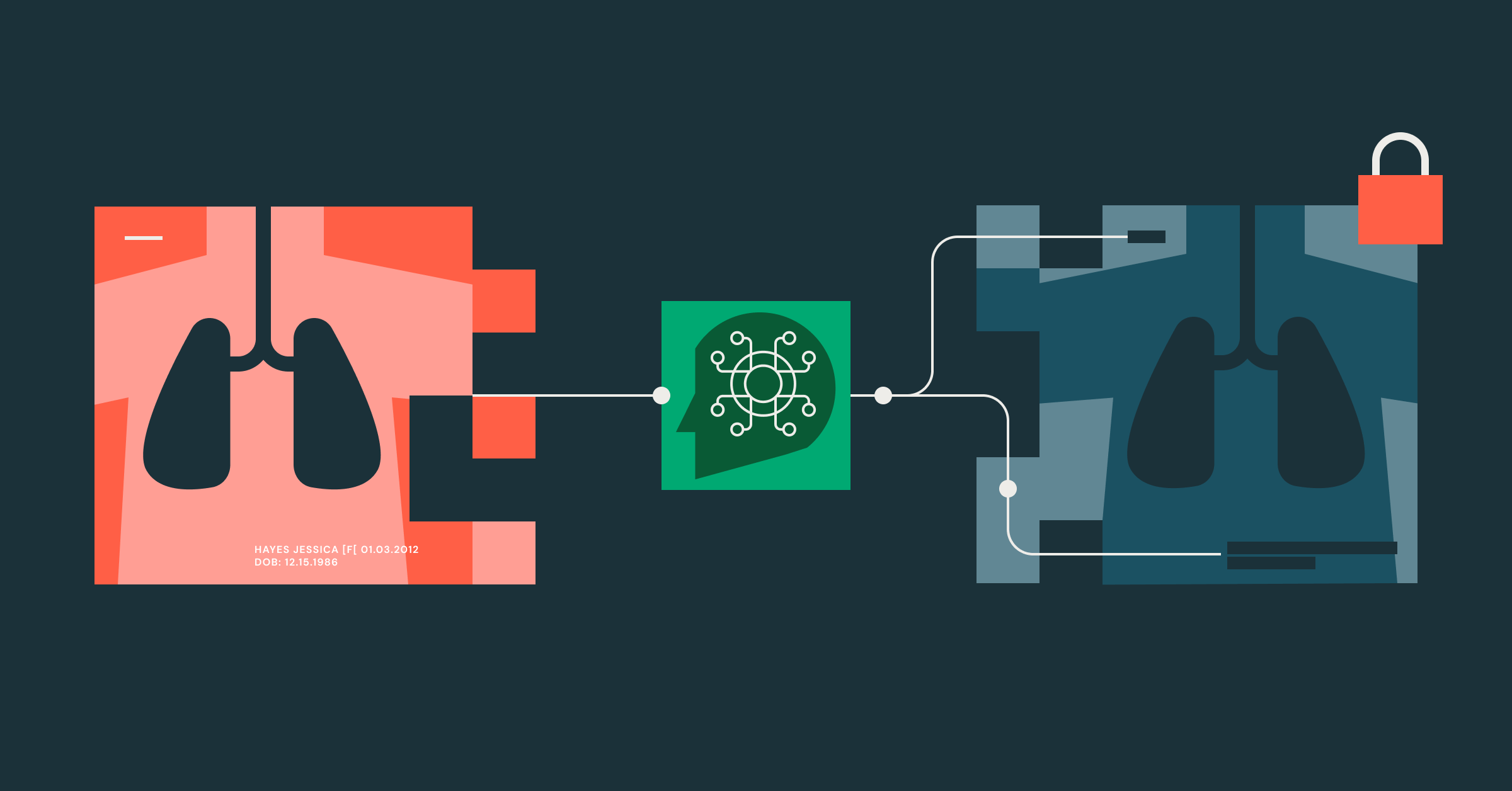
Summary
- Business Problem - To make medical records safe and useful for clinical research, and especially guard against unblinding a clinical trial, they need to be de-identified. While de-identifying text is a well solved problem, de-identifying images remains to be desired. We posit that VLMs have advanced to the point of becoming a viable solution for large-scale image de-identification.
- Solutions tested - During the course of this project we tested Presidio, an open source “go-to” solution, and commercial VLMs and open source VLMs (Visual Language Models)
- Results - In our experiments, VLMs are surprisingly good at detecting PHI in an image, with a recall of 100%. Besides their superior performance, VLMs are cost-effective and easy to use with little tuning compared to classic OCR and text-only approaches.
Why the need for scalable image de-identification
Medical images, such as X-rays and MRIs, besides aiding in diagnosis, treatment planning, and disease monitoring, are increasingly being used beyond individual patient care to inform broader medical research, public health policy, and the development of new AI-powered diagnostic tools. This secondary use of medical records, while immensely beneficial, needs to undergo de-identification of protected health information (PHI) to safeguard patient privacy and comply with regulations like HIPAA.
The growing scale of medical image datasets necessitates reliable and efficient de-identification methods, ensuring that the images can be safely and ethically used to advance medical science. To this end, we present the Pixels Solution Accelerator with a Spark ML Pipeline leveraging Vision Language Models (VLM) in parallel to de-identify medical images in the widely used format, Digital Imaging and Communications in Medicine (DICOM).
A DICOM file contains both images and metadata text (read more here). Here, we focus on our new feature de-identifying images. It is worth noting that Pixels, our DICOM toolkit, also de-identifies metadata in addition to scalable DICOM ingestion, segmentation, all within an web application.
How to de-identify PHI burned in DICOM images
After installing the Pixels python package, run the DicomPhiPipeline as such:
It reads in a DICOM file path in a column in a Spark dataframe and outputs 2 columns:
- a response from a VLM (specified in endpoint)
- a DICOM file path with PHI masked
As part of DicomPhiPipeline, redaction is performed using EasyOCR. Redaction can be performed independently of VLM PHI detection (redact_even_if_undetected=True) or performed conditionally on VLM PHI detection (redact_even_if_undetected=False). We recommend the latter as EasyOCR has a tendency to over-redact non-PHI. Conditioning on images the VLM has detected as PHI-positive, EasyOCR will be less likely to redact the non-PHI images.
Data intelligence reshapes industries
Comparison with other PHI detection methods
The competition
We tested Pixels’ image PHI detection pipeline with a commercial vendor and a widely used open source solution, Presidio. Both the vendor and Presidio used OCR to first extract the text from the images and then apply a language model to classify if the text was PHI or not. The built-in OCR also segmented sensitive text and applied a fill mask within those bounding boxes.
Additionally, we compared several VLMs: GPT-4o, Claude 3.7 Sonnet, and open-source Llama 4 Maverick.
Datasets
The comparison was done on public DICOM datasets, MIDI-B where we downsampled to 70 images to create a balanced dataset with roughly equal number of images with PHI and without.
Results
| Task: PHI detection in DICOM images | MIDI-B (70) | ||||
|---|---|---|---|---|---|
| Solution | Cost Estimates per 100k images | Recall | Precision | Specificity | NPV |
| ISV (commercial) | $4,400 per month prepaid | 1.0 | 0.71 | 0.93 | 1.0 |
| Presidio (OSS) | $0 | 0.7 | 0.7 | 0.95 | 0.95 |
| Claude 3.7 Sonnet | $270 | 1.0 | 1.0 | 1.0 | 1.0 |
| GPT-4o | $150 | 1.0 | 1.0 | 1.0 | 1.0 |
| Llama 4 Maverick (OSS) | $45 | 1.0 | 0.91 | 0.98 | 1.0 |
Both Claude 3.7 Sonnet and GPT-4o had perfect PHI detection performance. Llama 4 Maverick had 100% recall but 91% precision as it sometimes mis-identifies non-PHI text on the image as PHI. Nevertheless, Llama 4-Maverick still provides good performance especially for users who lean towards over-redaction to avoid missing any PHI. In such a case, it has zero false omission rate of PHI (i.e. NPV close to 1) and recall of 1 so it may be a good balance between performance and cost.
In our tests, we used Presidio and the commercial solution out-of-the-box with default settings. We noticed that performance in terms of both accuracy and speed was highly dependent on the OCR choice. It is likely their performance could be improved with alternatives such as Azure Document Intelligence.
Why it works
We surveyed the literature on de-identifying burn-in text on medical images and learned from the reported success of using OCR, LLMs (e.g. BERT, Bi-LSTM, GPT) and/or VLMs. Our decision to use VLM to detect PHI and EasyOCR to detect text bounding boxes was guided by the success reported by Truong et al. 2025.
VLM replace traditional OCR poor at text recognition and often introduce typos
In most de-identification methods reported, OCR was often used as the first step to extract text from images input into a LLM. However, we observed that OCR tools like tessaract and EasyOCR were generally poor and slow at text recognition (i.e. reading), often mis-reading certain characters and inadvertently introducing typos and compromising downstream PHI detection. To mitigate this, we used a VLM to read burn-in text and classify if the text was PHI; the VLMs were surprisingly good at this.
EasyOCR to detect bounding boxes for redaction when VLMs cannot alter images
However, VLMs cannot output redacted images. Thus, we used OCR to do what it did best, i.e. detect text, to provide the bounding box coordinates for subsequent masking. It is worth noting that although there have been recent attempts to fine-tune a VLM to output bounding box coordinates Chen et al. 2025, we opted for a simpler solution assembling off-the-shelf tools (VLM, EasyOCR) instead.
Spark parallelism for production-grade scalability
While Databricks had a batch inferencing capability with LLMs (ai_functions), it currently lacks support for VLMs. As such, we implemented a scalable version for VLM and EasyOCR using Pandas UDF. Working with a large pharmaceutical customer, Spark parallelism sped up their de-identification process from 105 minutes to 6 minutes for a trial run of 1000 DICOM frames! Scaling up to their full workload of 100,000 DICOM frames, the speed up and cost savings were significant.
Summary
Given the power, ease and economics of VLMs as demonstrated by the Pixels 2.0 solution accelerator add-ons, it’s not only feasible but prudent to protect your critical clinical studies and related image studies with scalable PHI detection.
While Pixels is designed for DICOM files, we found our customers adapting it for other image formats in JPEG, Whole Slide Images, SVS and so on.
The updates are posted to our github repo so now is a good time to update or try out the Databricks Pixels 2.0 solution accelerator. Reach out to your Databricks account team to discuss your imaging data processing and AI/ML use cases. The authors would be happy to hear from you over LinkedIn if we haven’t already been introduced.
Never miss a Databricks post
Sign up
What's next?

Data Science and ML
November 14, 2024/2 min read
Providence Health: Scaling ML/AI Projects with Databricks Mosaic AI
Product
November 27, 2024/6 min read