Genesis Workbench: A Blueprint for Life Sciences Applications on Databricks
Summary
- Transformative Potential: Generative AI is revolutionizing biotech R&D by utilizing foundation models to drive advancements in drug discovery, predictive modeling, and personalized medicine.
- Operational Barriers: Scientists currently face significant friction due to technical overhead, such as configuring GPU environments, managing complex workflows, and handling access controls.
- Streamlined Solution: Genesis Workbench addresses these challenges by providing a blueprint on Databricks to simplify the development of life sciences applications.
AI is Accelerating Target Discovery and Transforming Drug Design
The life sciences are experiencing a fundamental transformation, driven by the convergence of massive genomic and multi-omic datasets with advances in artificial intelligence (AI) and machine learning (ML). AI tools, initially specialized, have quickly evolved into sophisticated, generalized foundation models, demonstrated by innovations in protein structure prediction, protein language models, and even advanced generative models. These foundation models, trained on a vast corpus of biological sequence and structural data, are enabling accelerated drug discovery through processes such as lab-in-the-loop design, improving the efficiency of scientists' daily tasks, and potentially facilitating de novo design. This acceleration is further supported by specialized large-scale language models (LLMs), which are critical for the precise extraction and synthesis of biomedical knowledge from scientific literature, Electronic Health Records (EHRs), and Electronic Lab Notebooks (ELNs).
Practical challenges in building a secure and scalable research platform
Technologies to better find targets or design new drugs, as well as the data used in them, are highly protected Intellectual Property (IP). Organizations, therefore, need to ensure that appropriate access controls can be granted to individuals and groups, both for the data and the models. Additional governance concerns, such as auditing model use and cost monitoring, are key factors for foundation models on large-scale biological datasets. Importantly, however, these access control processes should not be so challenging to implement or so opaque to users that they hinder real progress by the scientific community within the organization. In practice, many organizations struggle to find this balance.
Despite their expertise in biology or computational biology, many highly talented scientists struggle to set up advanced biological models due to the burden of tasks related to niche aspects of modern AI technology. These challenges include technical complexities such as configuring CUDA environments for GPU acceleration, which is essential for efficiently training large models. Additionally, scientists often need to create and manage complex workflows that automate and efficiently scale data processing, model training, and MLOps. These tasks often require skills outside traditional biological training. Data engineering also poses a significant hurdle, involving the collection, cleaning, and integration of diverse biological datasets while ensuring compliance with data governance policies to maintain privacy and reproducibility. These non-biological demands divert valuable time and focus away from core scientific research, slowing progress and innovation when applying generative AI models in the life sciences. Addressing this gap requires interdisciplinary collaboration and improved access to tools that lower the technical barriers for biological researchers.
Genesis Workbench: A Blueprint for Biological AI/ML on Databricks
Effortlessly powerful: Databricks makes data and AI easy
Databricks stands out as a unified analytics platform that combines robust governance, intuitive usability, and comprehensive capabilities to build any data or AI solution you need. With centralized tools for data management, security, and compliance, it ensures your data is always protected while remaining easily accessible to users of all skill levels. Its seamless collaboration features, scalable processing power, and support for the full spectrum of data, analytics, and AI workloads make it the ideal foundation for organizations looking to innovate while retaining strict control and simplicity. Organizations both large and small have achieved success building biology models on Databricks, from Merck’s TEDDY Family of Foundation Models to Tahoe Therapeutics Single-Cell Atlas.
Genesis Workbench helps you supercharge life sciences on Databricks

Genesis Workbench provides a blueprint for developing Life Sciences applications leveraging Databricks capabilities. It provides working templates that utilize features such as automated workflows, GPU clusters, model serving, and MLflow to accelerate AI-driven life sciences research. It features an intuitive Databricks Apps interface with pre-packaged biological models and tailored workflows, enabling scientists to start quickly without complex setup.
In collaboration with NVIDIA, BioNeMo—a generative AI framework for digital biology—is integrated for easy access to advanced pre-trained models. The BioNeMo models are optimized for NVIDIA hardware, delivering high performance and scalability for enterprise workloads.

Being open source, Genesis Workbench provides extensible templates for AI engineers, reducing non-biological workload and promoting rapid innovation using and combining foundation AI models for biology. We provide a range of models with Genesis Workbench as a starting point, and these can be used by both bench scientists via the app, and by advanced computational users building pipelines. Importantly, through serving models on APIs one abstracts out the complex model dependencies and GPU requirements. This allows users to stitch together commonly used, but highly complex, tools in single pipelines.
Genesis workbench is a Databricks Solution Accelerator which is in active development and we expect our feature set to continue growing.
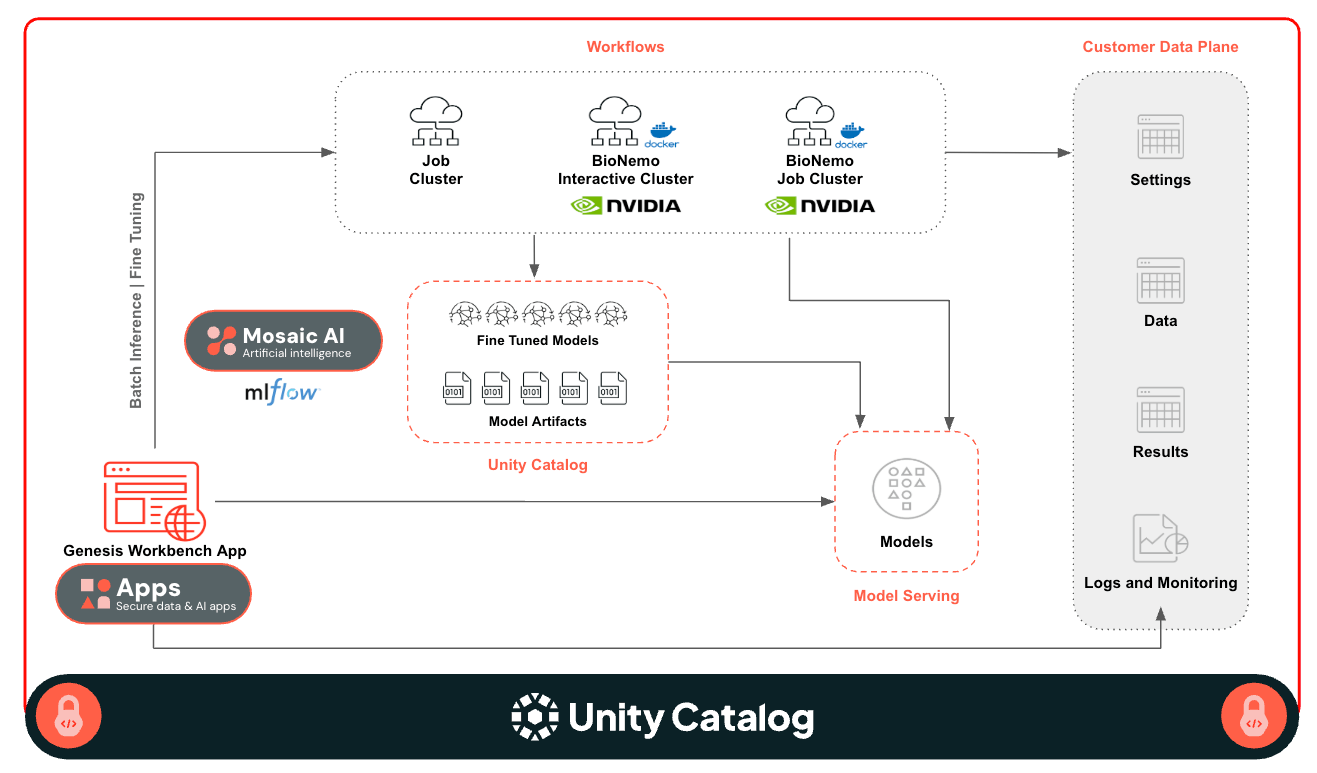
Genesis Workbench Architecture
This application is built on the Databricks platform, and it uses the following platform capabilities
- Unity Catalog for Governance
- Databricks Apps for UI
- GPU Model Serving for serving foundation models
- Lakeflow Jobs for running scalable batch inference and fine tuning
- Classic Compute with GPUs for interactive and batch workload
- Docker Container Service for integration of third party libraries
- Databricks Asset Bundle for easy deployment
Modules in Genesis Workbench
Protein folding and design
Overview
The ability to computationally predict the three-dimensional structure of a protein from its amino acid sequence has been a long-standing computational and theoretical problem. The CASP competition has been a venue where the latest advances have been tested against one another, furthering the research community’s modeling capabilities. DeepMind, with their initial alphafold model (Senior et al. 2020, Nature) and later alphafold2 (Jumper et al. 2021, Nature) releases in the CASP competition, were able to substantially improve on the performance of structure prediction. To do so, they utilized AI techniques to learn from previously measured structures and incorporate information from large sequence databases. The ability to predict many structures with near-experimental accuracy, including protein multimers, has revolutionized the approach to drug discovery.
There is now a zoo of models for protein structure-related tasks. Models such as Alphafold-3 (Abramsom et al. 2024 Nature)(closed weights), Boltz-1 (Wohlend 2024, BioRxiv), and Chai-1 (Chai discovery team 2024, BioRxiv), now extend beyond proteins to more advanced structures involving proteins, DNA, RNA, and small molecules. Other open models, such as Openfold3 (The Openfold3 team) have been recently released with similar capabilities, and this zoo of models is expected to continue growing. Additionally, the ability to generate proteins with generative models is rapidly advancing; tools such as RFdiffusion (Watson et al. 2023, Nature) and ProteinMPNN (Dauparas et al. 2022, Science) are often used in this space (see e.g. Bielska et al. 2025, front.immunol.), and new models such as BoltzGen (Stark et al. preprint, repo) and others provide a rapidly evolving toolkit for computational drug discovery and design
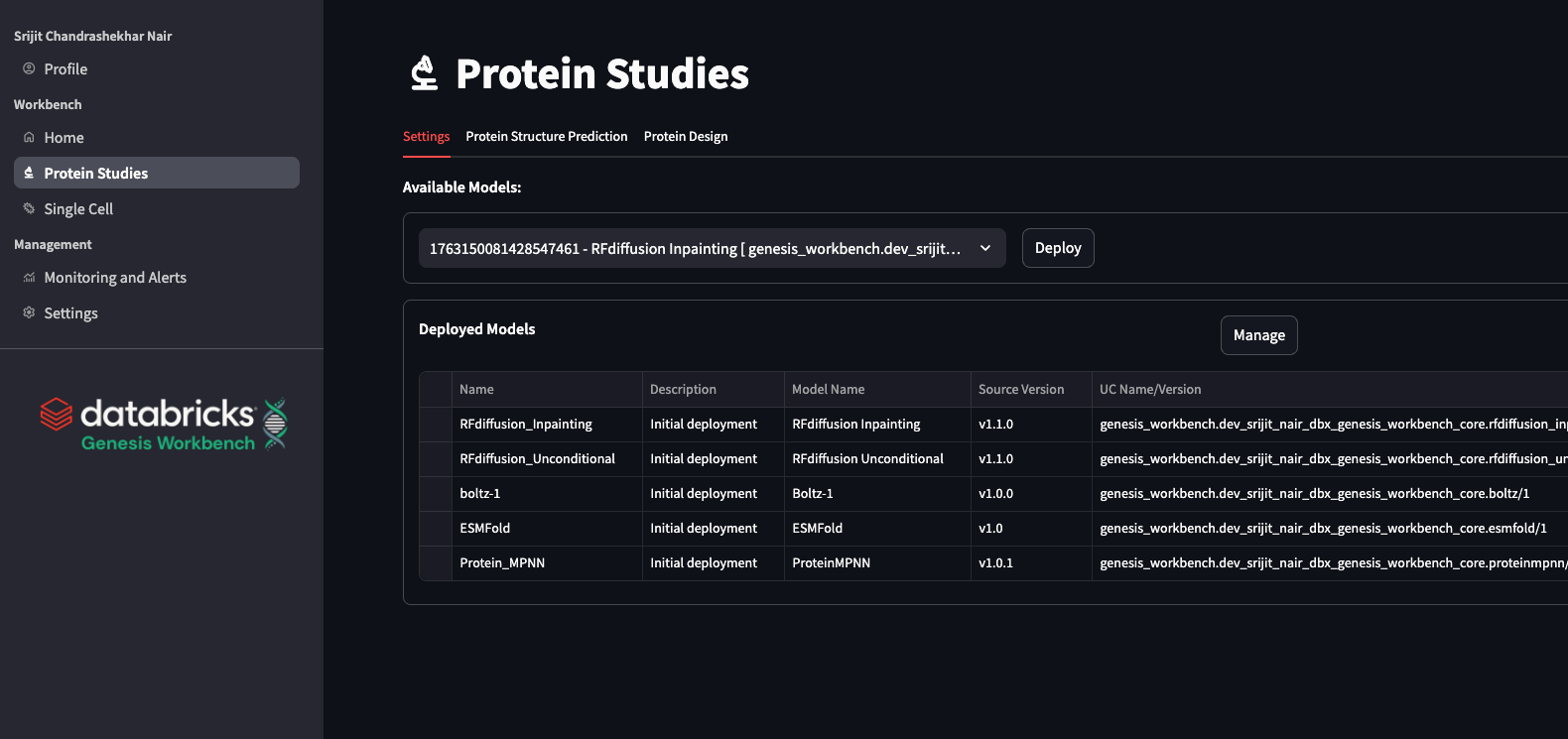
Alphafold
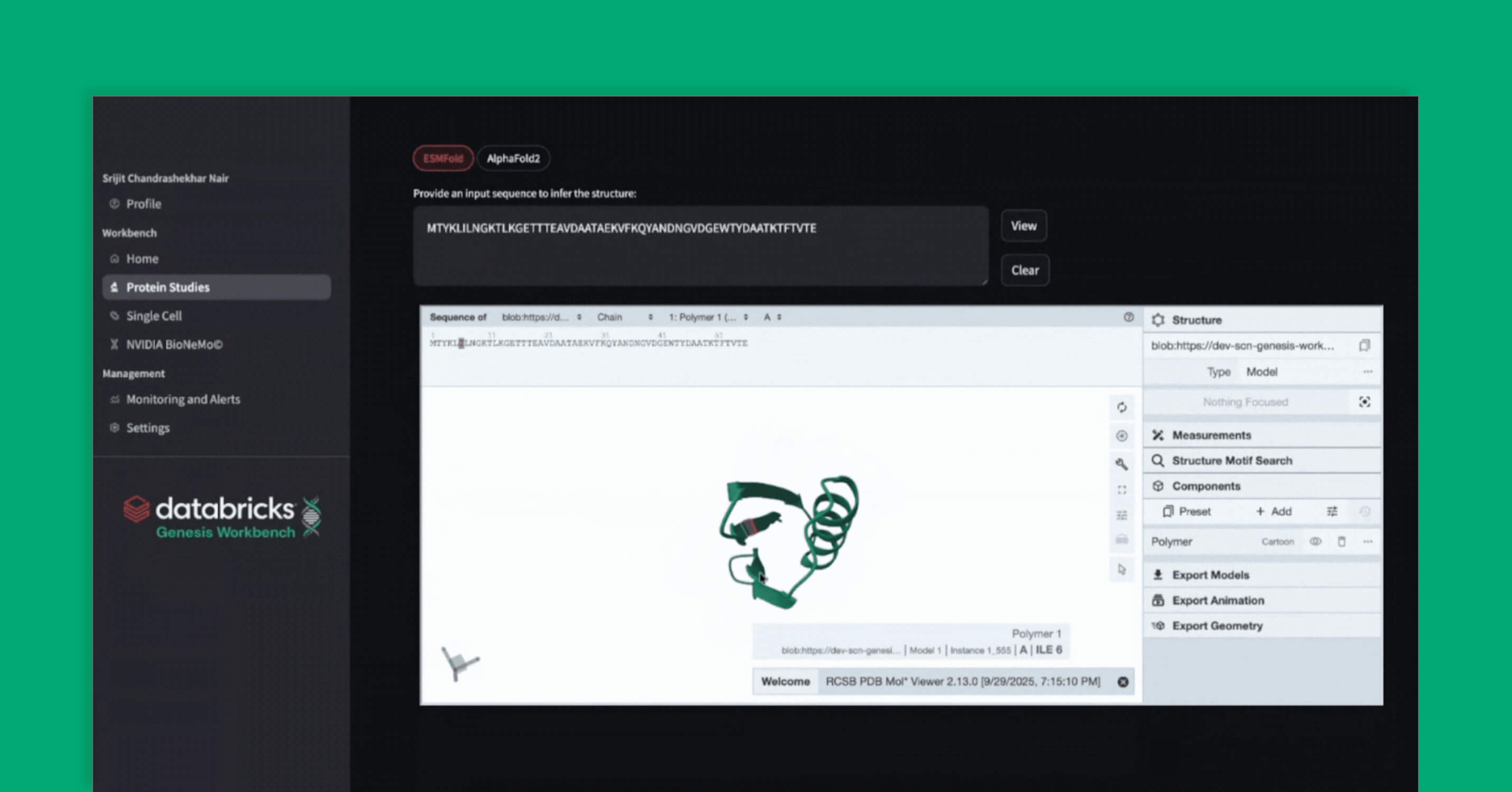
We include the latest stable version (v2.3.2) of alphafold2 (Jumper et al. 2021, Nature). Since the alphafold process takes some time to run due to the extensive feature computation, in Genesis Workbench, we show you how to provision alphafold as a Workflow Job in Databricks. The job is composed of two tasks: the CPU-dominated task (MSA, feature extraction) and the GPU-dominated folding task. This allows us to run each task on the appropriate compute type. The job utilizes a queueing system, much like an HPC, allowing for multiple sequences to be folded simultaneously while others wait in the queue.
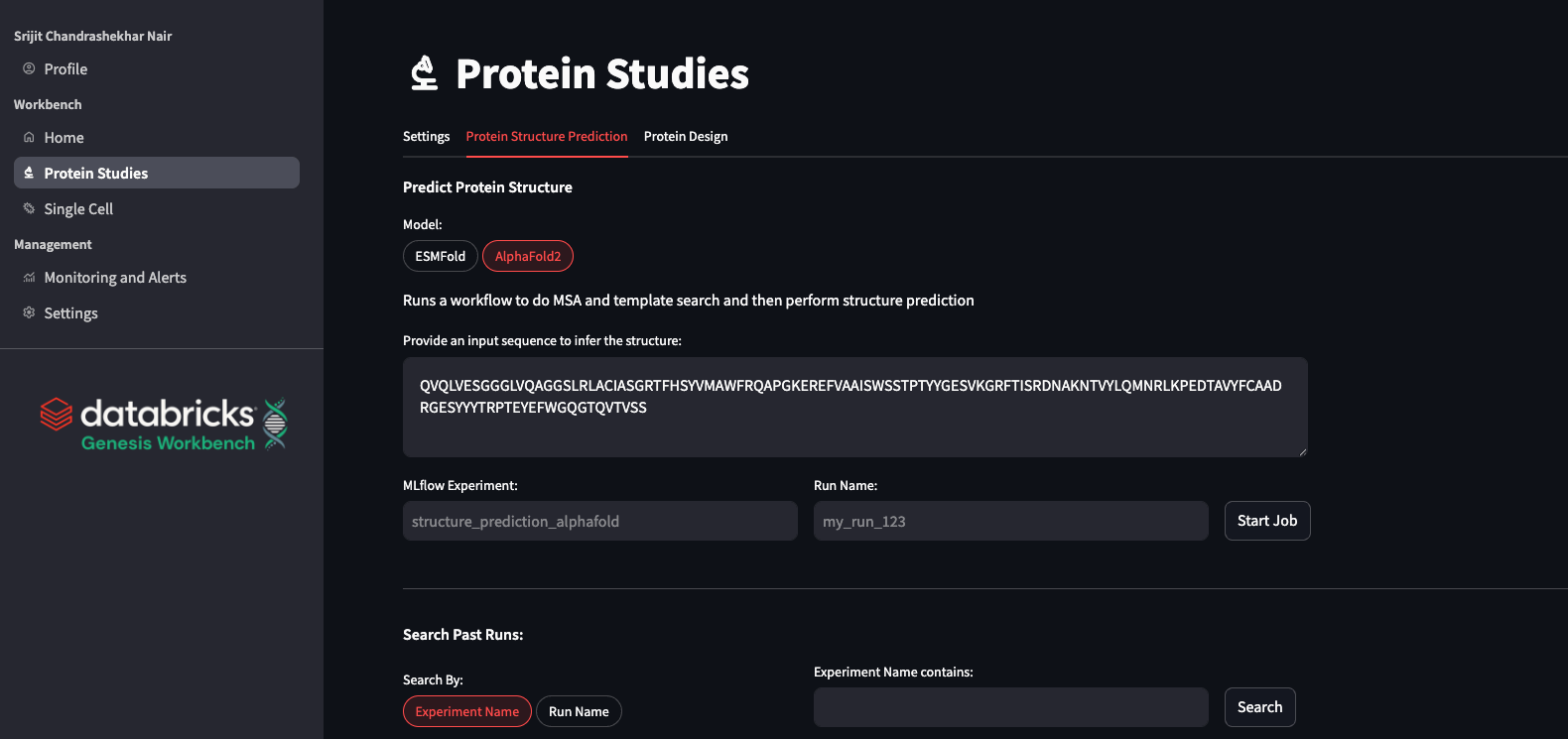
ESMFold
ESMFold (Lin et al. 2023, Science) is a fast, cost-effective, deep learning model for protein structure prediction that leverages powerful ESM protein language models and a structural head to infer protein structures from a single sequence. While its accuracy is lower than models using multiple sequence alignments, ESMFold is sufficient for many practical applications. In Genesis Workbench, we deploy ESMfold, wrapped in an MLflow model, and register it in Unity Catalog. Serving a Unity Catalog-registered MLflow model as an API is made extremely simple in Databricks, including support for scale-to-zero operations for cost savings. With ESMFold served as an API, researchers can predict new protein structures quickly, integrate it easily into computational pipelines, and visualize or download results for further analysis using patterns shown in Genesis Workbench.
Boltz
Boltz-1 (Wohlend 2024, BioRxiv) provides a fully open source code and weights (MIT) model for biomolecular structure prediction. Like ESMFold, we register Boltz-1 as a model in the Unity Catalog and serve it on a model-serving endpoint. Boltz-1 has an optional MSA input; one can omit it, use a precomputed MSA, or use an mmseqs2 (Steinegger, Sölding 2017 Nat.Biotech) server address. To ensure a range of options for Boltz-1, we wrap the essential Boltz-1 model components as a Python package together with JackHMMer (Johnson et al. 2010 Bioinformatics) options from the alphafold2 (Jumper et al. 2021, Nature) library. This allows users to choose JackHMMer MSA, which can be advantageous for some users since the default mmseqs2 server address is a public URL, which breaks most security policies.
Boltz-1 functions and MSA stages are wrapped with mlflow tracing. Tracing allows one to easily visualize all stages in the inference, and the input and outputs from each stage, for instance, for the MSA stage. This can be particularly useful when using the model in a notebook, as it allows one to quickly see exactly what happened in the model and identify issues with the MSA without needing to navigate to any stored files separately.
Protein Design with ProteinMPNN and RFDiffusion
Computational protein design is a rapidly evolving space with great potential for changing how we design effective proteins for both enzymatic activity and therapeutic effects. RFdiffusion (Watson et al. 2023, Nature) and ProteinMPNN (Dauparas et al. 2022, Science) are often used together in protein design, for instance, in computational antibody design (Bielska et al. 2025, front.immunol.) and recently even for de novo antibody design (Bennett et al. 2025, Nature).
In Genesis Workbench, we demonstrate how to serve RFDiffusion and ProteinMPNN. We demonstrate how, once the complex dependencies are abstracted in the model serving, it is easy to stitch these models together into computational pipelines. In particular, we combine ESMFold, RFdiffusion, and ProteinMPNN, and call the entire pipeline from a small CPU-only machine hosting the Genesis Workbench application. We no longer need to maintain all dependencies of each model in a single place, or worry about conflicting CUDA versions. In the app, we also structurally align the outputs with the original predicted structure to display the original and designed sequences in the Mol* viewer.
This demonstrates how computational users can build new tooling based on the components within Genesis Workbench to create processes tailored to the goals of their organization or research team.
Single Cell analysis
Overview
Single-cell transcriptomics is a powerful technology for understanding cell populations across different cohorts, identifying niche cell types, elucidating cell trajectories, and much more. The scale of available data in this space is rapidly growing, not only through increased sample processing but also through improvements in technologies. This presents a challenge for data labeling and processing at this scale, raising the questions:
- Can some portion of the analysis be automated to prevent overburdening advanced PhD-level researchers by requiring them to spend time on repetitive tasks?
- How can we pre-annotate data to enhance data discovery for future atlas-level analyses of the samples?
We aim to provide solutions to these challenges and build on our blueprint for adapting these approaches to variations of these problems.
In Genesis Workbench, we provide tooling to automate single-cell processing and visualize the results, all tracked in MLflow experiments, so that users retain control over the data and how they share it with their peers. This includes CPU pipelines with optimal RAM selection and GPU-accelerated workflows. Further, we include responsive low-latency data visualizations of the processed data. We also incorporate foundation models of single-cell expression: SCimilarity (Heimberg et al., 2024, Nature) and scGPT (Cui et al., 2024, Nature Methods); for atlas-level annotation, embedding, and search.
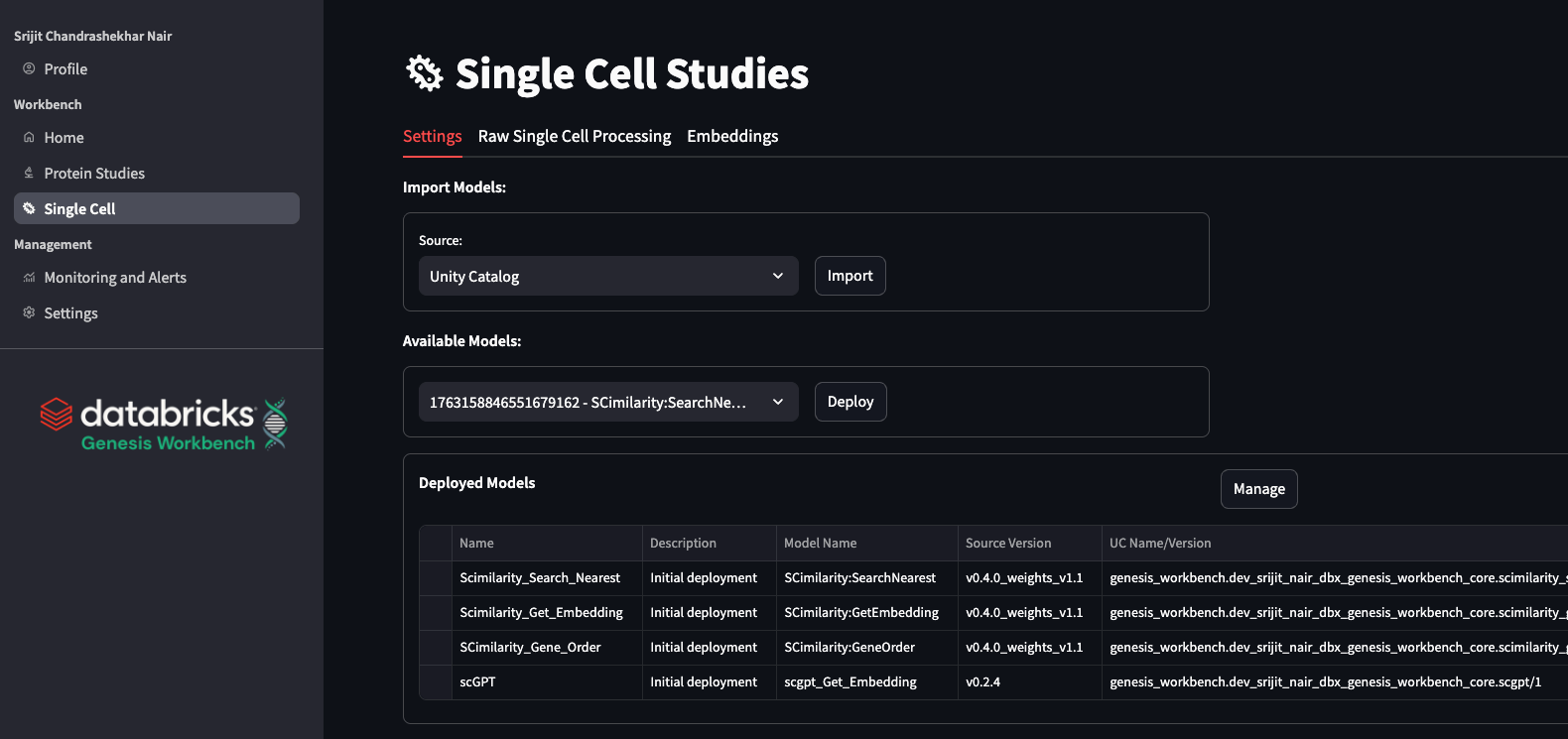
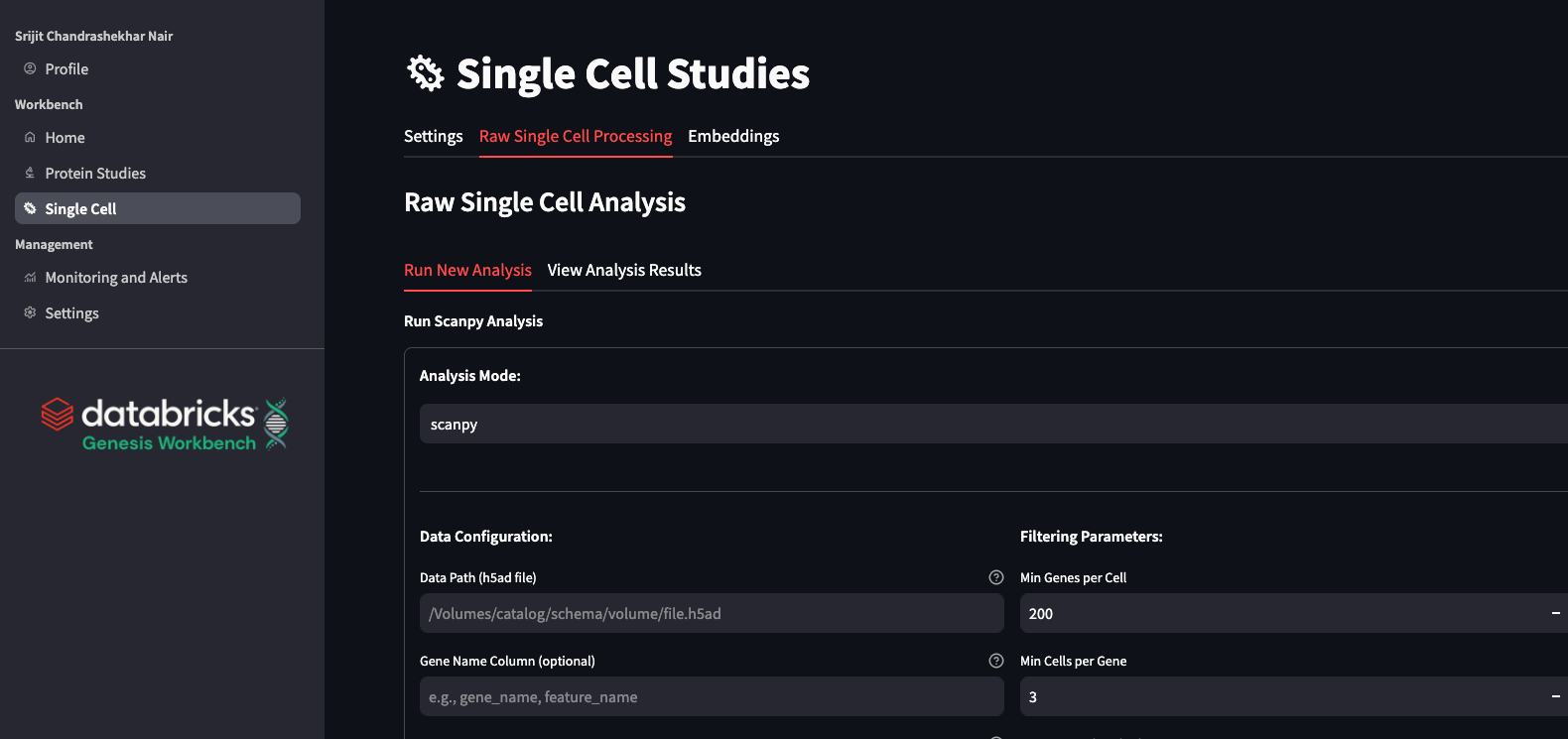
scRNA Data Processing: scanpy and rapids-singlecell
In Genesis Workbench, we construct a workflow job for a standard scanpy (Wolf et al. 2018 Genome Biol) run, which attempts to automatically select the smallest RAM size to lower user burden while simultaneously lowering cost. The job logs the chosen parameters as well as various metrics, outputs, and figures into a single MLflow run. This enables a modern experiment tracking procedure, with automated logging of all details regarding the pipeline being run. In addition to this Scanpy job, we also provision a RAPIDS-singlecell (Dicks et al., 2022, repo, doi) job. This package behaves very similarly to scanpy and has the same parameter options, but is accelerated by a GPU. This enables faster throughput, which is essential for certain projects.
In the Genesis Workbench application, we make these pipelines easily accessible to users via a simple interface that allows them to start runs with different parameters and view the output within the application. We perform intelligent data curation to enable a rapid in-app analysis of results, supporting multiple users on a single, compact application compute. Details such as QC metrics, clustering, and marker gene expression can then all be viewed in-app. This processing can be an important step before using data with foundation models for scRNA.
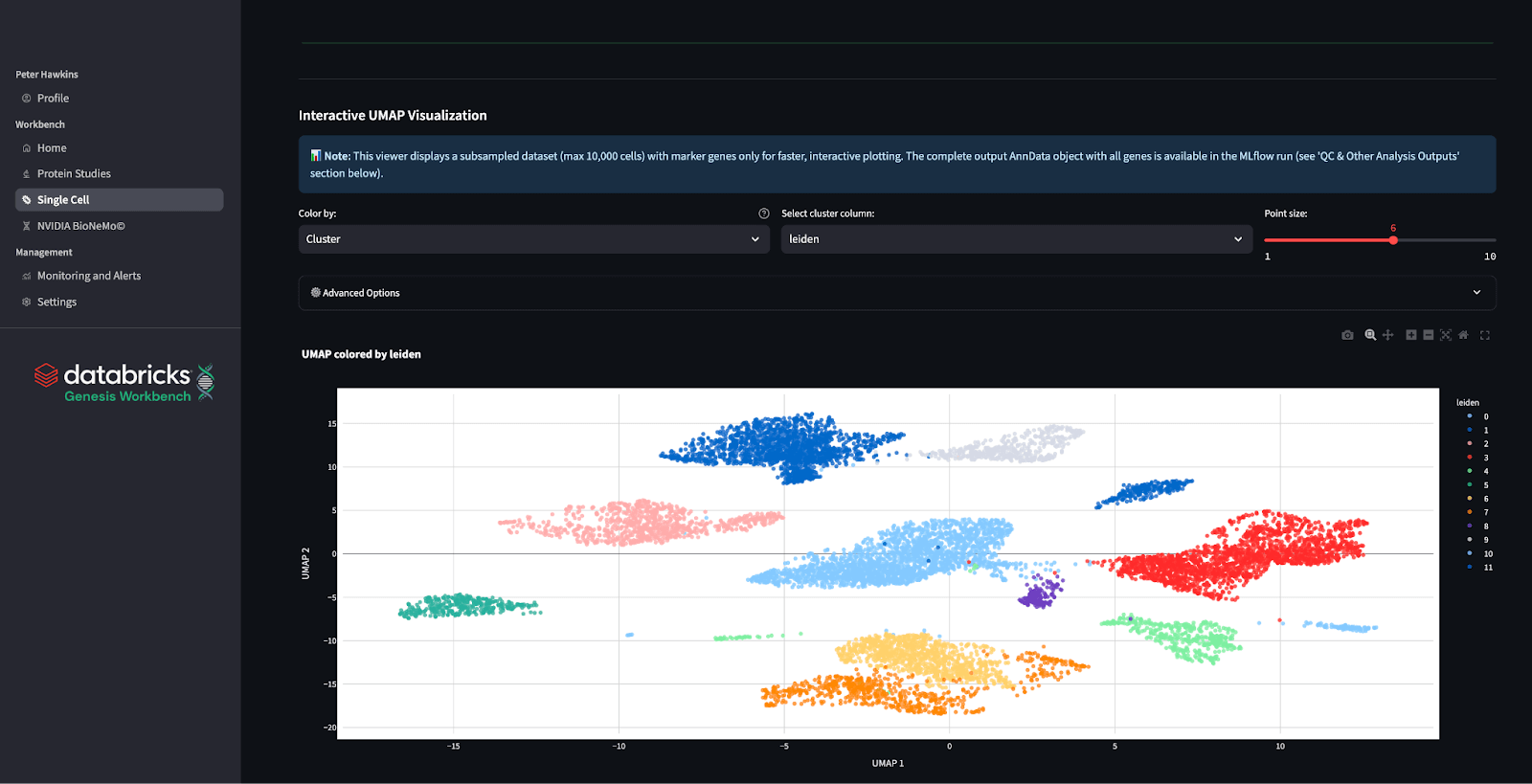
Foundation Models: SCimilarity and scGPT
The single-cell analysis module within Genesis Workbench provides life sciences researchers with access to two state-of-the-art foundation models—SCimilarity and scGPT.
SCimilarity utilizes metric learning to search a large-scale cell atlas for the rapid identification of disease-relevant populations and analogous in vitro models, achieving organ- and disease-level precision using reference catalogs without requiring clustering. Complementing this, scGPT employs a 53M-parameter transformer architecture with dual gene-cell embeddings for perturbation prediction, regulatory network inference, and zero-shot multiomics integration. Together, they harmonize diverse datasets, minimize annotation bias, and reveal mechanistic disease insights—SCimilarity via Integrated Gradients and scGPT via attention mechanisms—driving translational discovery and therapeutic innovation across oncology, immunology, and rare disease research (Heimberg et al., 2024, Nature; Cui et al., 2024, Nature Methods).
Genesis Workbench deploys key endpoints for both models by packaging these foundational models with pre-configured GPU clusters, automated workflows, and MLflow integration: cell_query.gene_order, cell_embedding.get_embeddings, and cell_query.search_nearest for SCimilarity; gene embedding inference for scGPT. The example notebook demonstrates workflow based on the SCimilarity’s IPF myofibroblast tutorial — loading and normalizing scRNA-seq data, computing embeddings, and querying 23.4M cells to identify nearest disease-enriched populations using the served endpoints. Identified populations will enable downstream scGPT analysis for gene regulatory inference, perturbation screening, and CRISPR predictions within a single environment.
Databricks and NVIDIA Deliver Speed and Scale
NVIDIA’s software stack offers powerful solutions for accelerated life sciences research. NVIDIA BioNeMo is an open source framework that accelerates the development of deep learning models for biopharma. It enables researchers to scale biomolecular AI models, spanning DNA, RNA, and protein data, to new heights with rapid, streamlined tools. Parabricks delivers rapid, high-throughput genomic analysis using GPU-optimized algorithms, enabling swift processing of next-generation sequencing data for genomics labs. Rapids-SingleCell enhances single-cell data analytics, allowing scalable, interactive exploration and analysis of large multi-omic datasets by leveraging GPU acceleration for deeper biological insights.
The close collaboration between NVIDIA and Databricks engineering teams enabled seamless integration of NVIDIA’s advanced software stack into Genesis Workbench, delivering rapid and scalable AI-powered workflows for life sciences. Using patterns in Genesis Workbench, you can:
- Leverage BioNeMo models on Scalable GPU Compute inside the Databricks Platform
- Fine-tune models available in the BioNeMo package and use them for inference at scale
- Use RAPIDS-singlecell, an open source, GPU-accelerated framework for single-cell analysis
- Use parabricks for genomics analysis
Streamlined Visibility and Cost Optimization Using Databricks Dashboards
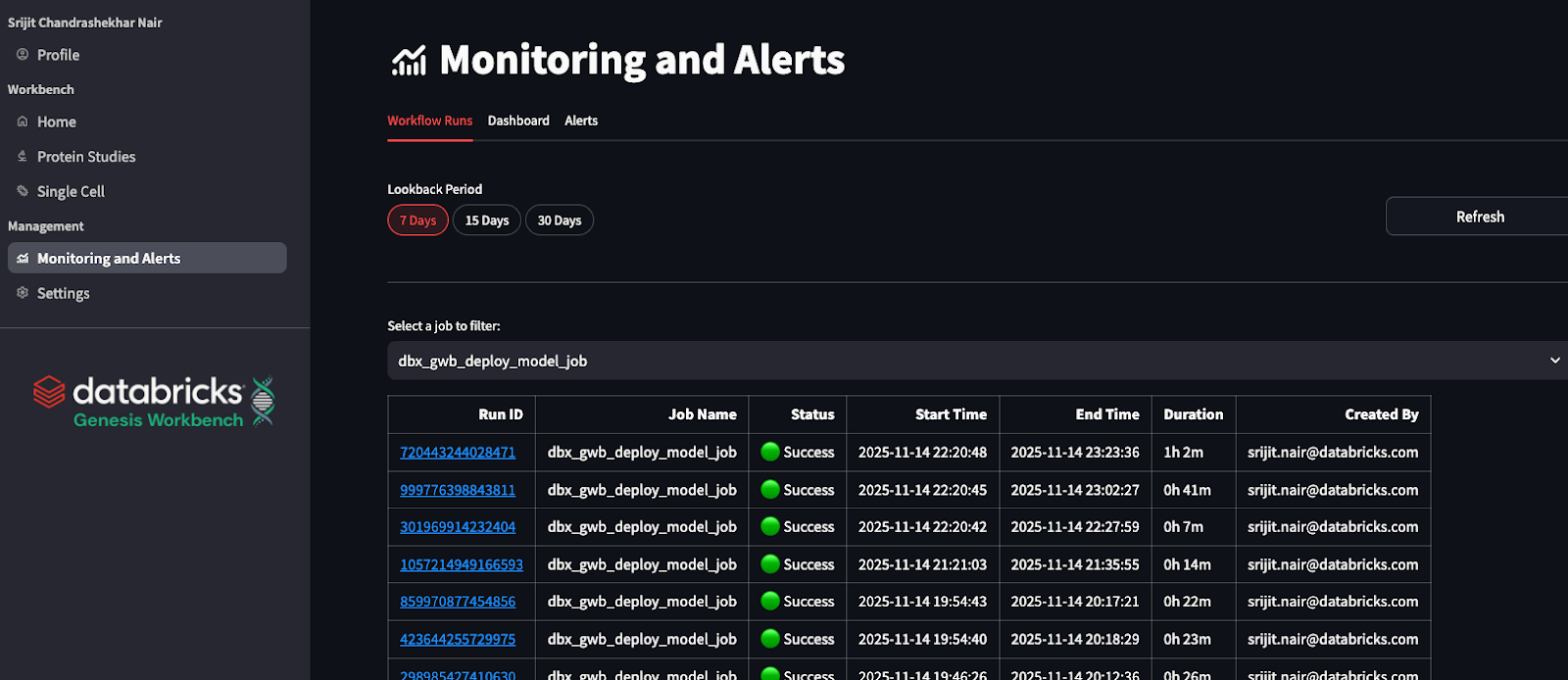
Monitoring Job Runs
Few workflows require long-running background processes, and the application leverages Databricks Workflow and launches jobs asynchronously. Genesis Workbench includes a dedicated monitoring dashboard that displays all Databricks Workflow jobs launched from within the system.
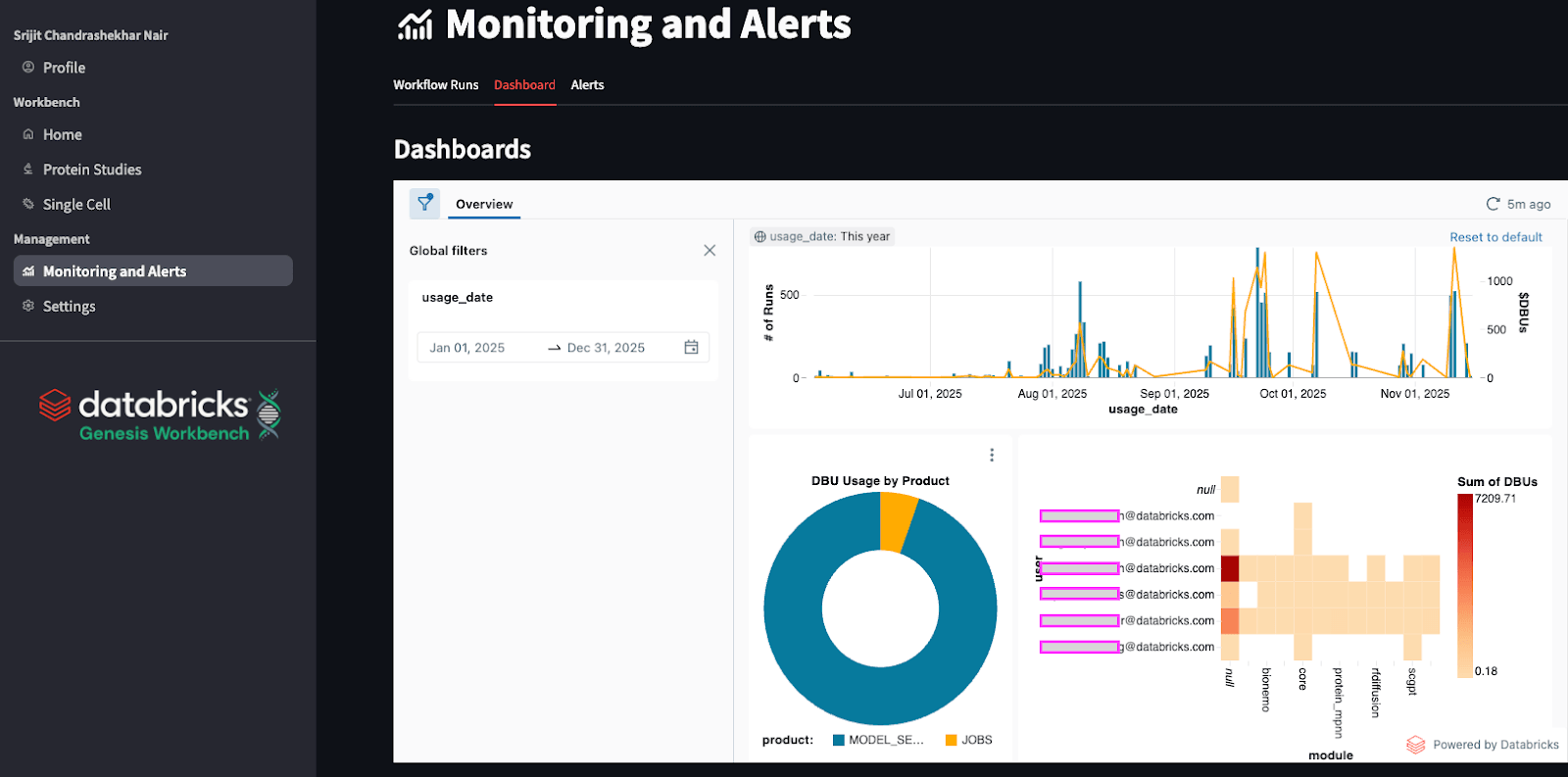
Cost Monitoring
Building computational pipelines and tooling with foundation models often raises questions about the associated costs. Databricks captures information about usage and billing automatically in system tables. This means one can easily build dashboards over this information to create custom ways to quickly view and analyze usage and spend. One can also use these to create automated alerts relating to overuse. In Genesis Workbench, we automatically provide a dashboard within the application that shows the spend on each model, broken down by time and user. Additionally, the dashboard's format is also open source and can be customized by teams that install Genesis Workbench.
Data intelligence reshapes industries
Next Steps
Installation
Genesis Workbench includes a script that utilizes Databricks Asset Bundles to deploy the application. The application can be downloaded and installed from the GitHub repository by following the installation instructions provided.
The source contains:
- scripts to deploy the Genesis Workbench core module in your workspace that includes the UI application
- scripts to deploy below modules:
- Single Cell module
- Protein Studies module
- BioNeMo module that contains container definitions and workflows (NOTE: The BioNeMo container needs to be built separately)
- Monitoring and Dashboards
Feedback
This project is currently in active development, and we look forward to hearing from you. Please reach out to your account team to discuss your use case and understand how you can leverage the patterns discussed in Genesis Workbench.
Roadmap
- More models provided out of the box
- Advanced MSA support for faster protein folding with MSAs
- Streaming support for single cell data
- BioNemo integration with model serving
- Spatial transcriptomics support
Conclusions
Genesis Workbench offers out-of-the-box MLflow packaged biological foundation models and NVIDIA BioNemo models, which integrate seamlessly with the Databricks ecosystem. This is a blueprint for how Databricks users can bring new models into Databricks, combine multiple models together into complex pipelines, and support both bench and computational scientists with a single framework.
Databricks’ governance via Unity Catalog provides a consistent, overarching system for controlling access to models, jobs, data, and the application. This can dramatically simplify the development of applications and data pipelines using these foundation models while ensuring appropriate access control.
Databricks model serving capabilities allow computational scientists and MLOps engineers to abstract out the complex dependencies and GPU requirements of each model. This allows other computational users to build pipelines, stitching together multiple models with their own dependencies, without needing to worry about it. This dramatically increases the development lifecycle, and can be easily shared across teams with appropriate permissioning thanks to Unity Catalog.
Databricks Apps provides a fast developer life cycle for testing and building applications based on these foundation model building blocks and biological data viewers. Since the Genesis Workbench code is open-sourced, teams can adapt and refine the app to suit their teams’ needs. They can add new models as needed, as well as follow the blueprint provided by Genesis Workbench to build the system as required for their business.
Genesis Workbench is a Databricks Solution Accelerator in active development. We encourage teams interested in using this tool to engage with us, provide feedback, and help shape our future roadmap.
Never miss a Databricks post
What's next?

Data Science and ML
November 14, 2024/2 min read
Providence Health: Scaling ML/AI Projects with Databricks Mosaic AI
Product
November 27, 2024/6 min read