Introduction
Large Language Models (LLMs) have given us a way to generate text, extract information, and identify patterns in industries from healthcare to finance to retail. In order to produce accurate, non-biased outputs, LLMs need access to high-quality datasets. In short: reliable data produces robust models. Obtaining this high-quality data often entails processing vast amounts of raw information; speed and efficiency are crucial as text is filtered, tokenized, encoded, and converted to embeddings.
Databricks and MosaicML offer a powerful solution that makes it easy to process and stream data into LLM training workflows. Databricks delivers a world-class Apache Spark™ engine for data processing and a unified data governance solution known as Unity Catalog (UC). With MosaicML's tools, Databricks customers now have the opportunity to unlock the full potential of Spark for pre-training and fine-tuning LLMs on their own data.
Common data formats like JSON, Parquet, and CSV often fall short when it comes to LLM training. These formats lack compressibility and are not structured to offer fast random access, critically impacting performance and model quality (due to poor data shuffling). Plus, as data sets grow larger and larger, loading data into a training run becomes increasingly complicated. What if data is stored in multiple locations or is in a less-than-ideal format? To address these challenges, the MosaicML engineering team created the open source StreamingDataset library to make multi-node, distributed training as fast and easy as possible. StreamingDataset makes it simple and efficient to stream data from any source and supports a variety of formats, including CSV, TSV, JSONL, and our versatile Mosaic Data Shard (MDS) format.
MDS is a data format specifically designed for efficient training of Generative AI models. It offers a number of benefits over other formats, including:
- Performance: MDS provides high throughput data loading and high-quality shuffling due to extremely fast random access to training samples. Data is sharded and stored in a row-based format which enables prefetching of samples, while shard information metadata is stored in an index file. The header data inside the shard file enables fast lookups of the row location of a given sample.
- Scalability: MDS can be used to store and train very large data sets (even those with trillions of tokens) and scales with distributed training frameworks. As a dataset grows, it's easy to add more shards and distribute the load across multiple nodes.
- Flexibility: MDS can be used to store a variety of data types, including text, images, and audio.
To make MosaicML's LLM training infrastructure available to all Databricks customers, we added two critical components: the ability to transform raw data into the MDS format and a new UC Volume backend that supports remote usage of data shards by MosaicML's runtime engine. In this blog post, we'll explain how customers can leverage proprietary data stored in UC Volume to train custom LLMs with MosaicML's Streaming Dataset Library. Let's get started!
Convert Data to MDS Format using Spark
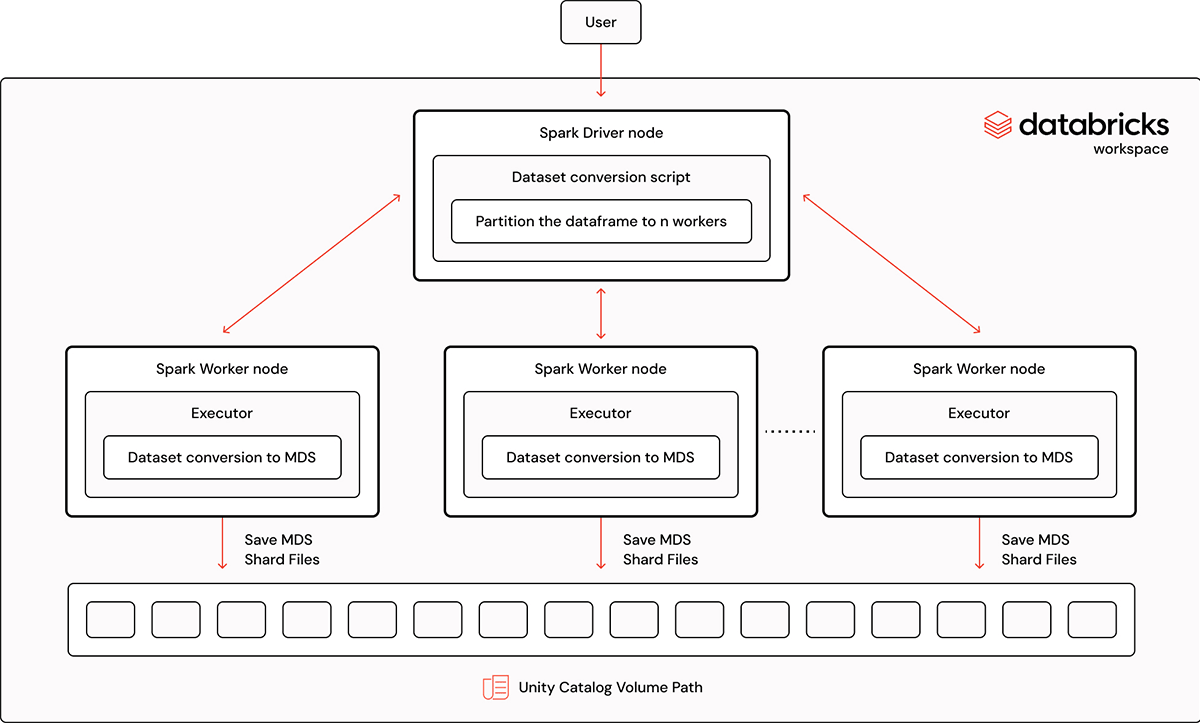
In this tutorial, we'll walk through the process of converting a Spark data frame into a StreamingDataset MDS format using our streaming Spark converter. This powerful tool allows users to efficiently convert a large data set into the MDS format. It also provides the option to chain the conversion with other preprocessing jobs like deduping or tokenization. This feature is particularly useful when materializing intermediate dataframes requires additional development effort or time. The complete example notebook can be found in MosaicML's Streaming library.
1. Installation of Libraries:
Ensure the necessary libraries are installed, including the mosaicml-streaming package and related dependencies. Use pip to install them as follows:
2. Basic Use-Case: Convert Spark Dataframe to MDS Format
In use cases where no extra Spark jobs are required, users can try out these steps to make the conversion.
First, identify the UC volume path to the dataset that is being converted to MDS and make sure the data set has a format supported by Spark. Then, load the dataset into a Spark dataframe using a specific Spark command.
Converting the Spark dataframe to MDS format takes one simple call from the Streaming library. In the below example, we supply the mds_kwargs to the function as it specifies the output folder location, which can be either a local directory, a remote cloud bucket, or both. It also specifies the schema of the input data frame. There is a schema sanity check implemented within the dataframeToMDS() function, so if the schema detected does not match the Spark data types, the user will know earlier and mitigate potential issues.
The data frame is divided into 4 partitions; in this example, each partition is converted into MDS format in parallel in a partitioned sub-directory. The number of partitions to use depends on several factors such as the size of the dataset and cluster. By default, each Spark task uses one CPU core. For maximum parallelism, the number of partitions can be set to the number of workers X the number of CPU cores per worker. However, this may not always be the optimal setting depending on the desired partition size; some user judgment is required.
The merge_index=True parameter merges the partitioned sub-directories dataset into one single data set. The duration of the function call depends on the size of the data frame and scales as the number of Spark workers increases, so users should configure their cluster accordingly. To give some practical examples: with a smaller dataset of 11GB with 5.8 million records, the MDS conversion finishes in 1 minute when using 8 workers with 32 cores each. For a larger dataset of 1.6T with 968 million records, we used 128 workers with 32 cores each and the MDS conversion finished in 5.6 minutes. We used Databricks' runtime 13.3x-cpu-ml-scala2.12 for all the experiments and all worker and driver nodes were i3.8xlarge with 244 GB memory and 32 cores.
3. Advanced Use-Case: Convert Spark Dataframe into Tokenized Format then Convert to MDS Format
In practice, users often want to chain tokenization and concatenation into one Spark job before materializing the data sets. This minimizes I/O operations and partition shuffling, which significantly speeds up the data processing pipelines. The MPT models were trained on a tokenized dataset to produce efficient, flexible, and generalized models. For those more complex scenarios, we provide an example below. We follow the same initial steps as in the basic use case but introduce additional tokenization for the data. All steps are the same as above, except that users can customize the iterator function by supplying a user-defined function that modifies the partitioned data frame. This function yields an output in dictionary format, with keys representing column names and values being the modified data (e.g., tokenized data). For this demonstration, the user-defined function is a simplified tokenizer. In practice, users can put in more complex logic processing for each record, as long as the function returns an iterable over the data frame.
After a user's own iterator function is defined, they can call the same API by supplying the callable function, as well as add extra arguments that the callable function may require. In this example, the udf_kwargs contains the configuration information for the tokenizer. The mds_kwargs is the argument for MDSWriter. The out parameter is the UC Volume path where MDS shard files will be saved.
Users can instantiate a StreamingDataset class by providing the converted data (shown above) with the following code, for immediate testing.
By following these steps, users can seamlessly convert Spark data frames into the StreamingDataset MDS format, enabling efficient data processing and analysis for various machine learning and data science tasks.
Streaming Data from UC Volume to MosaicML Platform
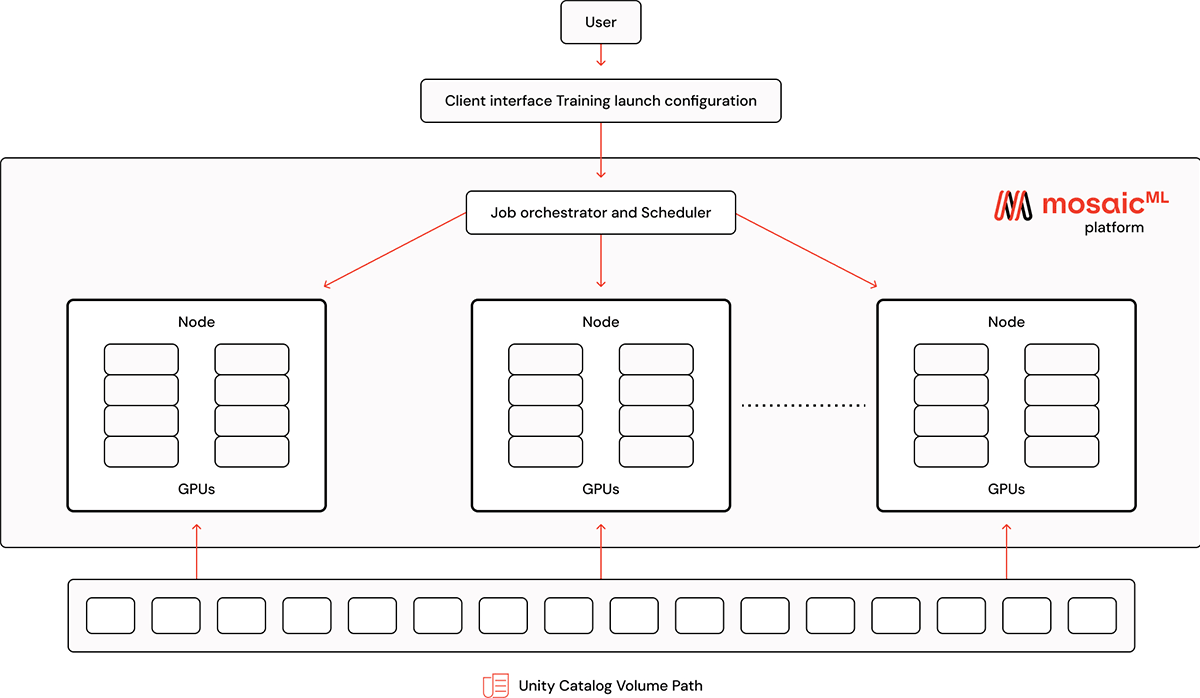
Now that our MDS files are stored in the UC Volume, we can use the dataset to train models on the MosaicML platform. To configure a training run, simply point a remote dataset to the UC Volume using the dbfs URI with the prefix dbfs:/Volumes
The volume path should point to the directory where the index.json file lives. This way, when fetching training data, MosaicML's platform can pull individual sample files from the directory.
Besides UC Volumes, the platform can be configured to stream data from DBFS using the same dbfs URI with the prefix as dbfs:
Credential Setup
Before starting to train on the MosaicML platform, it's necessary to configure authentication for the Databricks client. The simplest way to do this is using a Personal Access Token (PAT). For information on what a PAT is and how to create one, please see our documentation on authentication.
Once the PAT is created, the below MosaicML CLI commands are used to set the DATABRICKS_HOST and DATABRICKS_TOKEN environment variables. They should correspond to the workspace with access to the UC Volume that contains the MDS datasets.
See MosaicML CLI documentation on secrets for more information.
LLM Training
Once the above credentials are set, the MosaicML platform is ready to launch an LLM training job with data streaming from UC Volume. The MosaicML LLM Foundry contains code for training an LLM model using Composer, Streaming dataset, and the MosaicML platform. For demonstration purposes, we are using the mpt-125m.yaml with a tokenized Wikipedia dataset generated by dataframeToMDS API and materialized in the UC Volume path. Below is the sample MosaicML platform yaml:
An MCLI job template specifies a run name, a Docker image, a set of commands, and a compute cluster to run on.
Users can run the yaml on MosaicML platform using the command as follows:
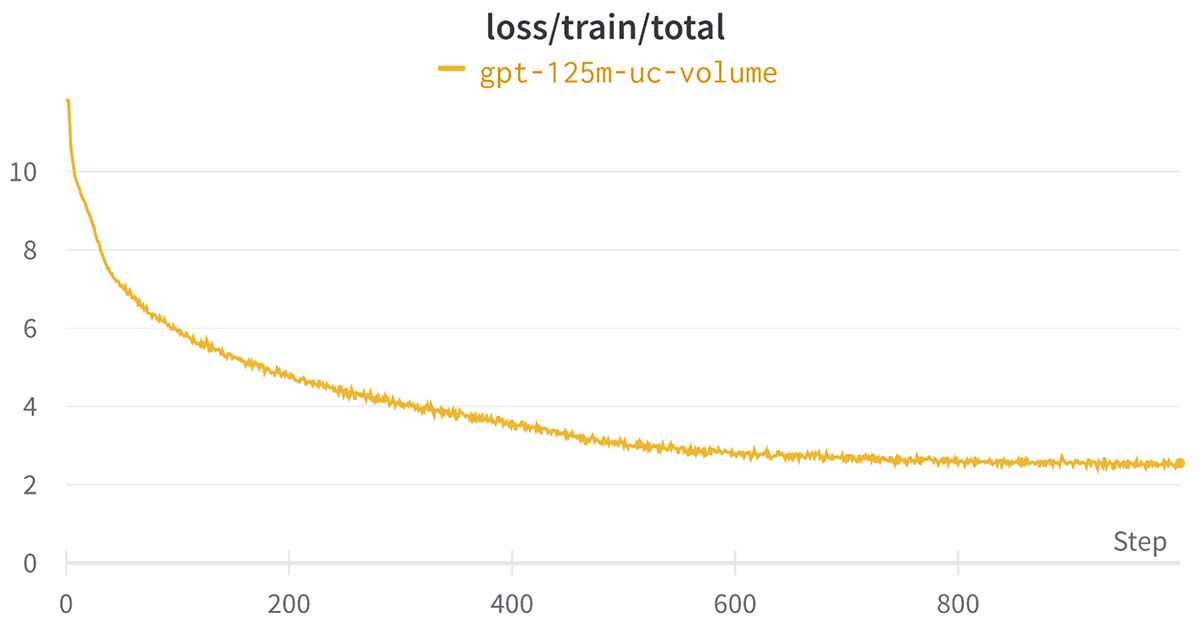
Below is the loss curve for the above job which shows a reduction in the loss as expected.
What's Next
In this blog post, we've taken the first step toward converting the data set to MDS in parallel across multiple workers and streaming data directly from the UC Volume. Next, we plan to improve the user experience of LLM training when data processing and streaming datasets directly from other data formats. Stay tuned for more updates! If you like MosaicML Streaming, give us a star on GitHub. Also, feel free to send us feedback through our Community Slack or by opening an issue on GitHub.
- Streaming dataset format documentation page.
- Dataset conversion to MDS format guide.
